Article Text
Abstract
Introduction Clinical trials generate a large volume of literature and a vast amount of data. Following the 'open science' model, data sharing has enormous potential to strengthen scientific research. Currently, to the best of our knowledge, there is no existing web based Hellenic biomedical registry that displays available patients for clinical trials, providing direct access to registered physicians to all data, assisting them in finding eligible patients in the initial clinical trial recruitment process.
Methods This paper describes the design and virtual implementation of a web based prototype biomedical registry in Greece. The system represents an eGovernment framework proposal for the central storage of patients' biomedical information and the operations associated with this process. The increasing tendency to include molecular data as prerequisite elements in clinical trials is adopted in the registry philosophy. The designed system is based on free, open source software and it is implemented virtually on a local host environment.
Results Using colorectal cancer as an example, valid data from patients increases the reliability index, demonstrating the functionality of the web application.
Conclusion In conclusion, the combination of biomedical data and information technology in order to display potential participants per health unit, facilitates recruitment for clinical trials.
- clinical trials
- recruitment system
- egovernment
- web-based framework
- colorectal cancer
- biomedical registry
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Summary box
What was previously known
A clinical trial is a demanding process and generates a large amount of data.
Finding eligible participants for a clinical trial is a challenging process.
Patient data sharing has great potential to strengthen scientific research.
What this paper adds
Develop an efficient web based framework to overview available patients per health unit and per International Classification of Diseases, 10th edition (ICD-10) codes, nationwide.
A doctor can find eligible patients based on their biomedical information.
Develop a potential patient recruitment system for all ICD-10 codes.
Promote medical collaboration.
Introduction
Clinical trials cover a wide range of different types of research in groups of participants for the evaluation of new drugs and vaccines, as well as new methods of treating and managing diseases, with the aim of answering questions and redefining considerations for new medical treatments. Each individual, in a normal or diseased state, has his or her own genome and his own environmental influences, resulting in a unique health record and an individual reaction to drugs. However, clinical trials often ignore this complexity of atomic information or attempt to include it only minimally, especially when focusing on small patient groups.1 Recruiting and grouping patients is an important process in the design of a clinical trial. Generally, the reliability of the research results is increased when standards are followed (including the specific disease of the patient population, similar approaches to treatment protocols, similar staging and specific age ranges), according to https://clinicaltrials.gov/ct2/about-studies/learn (last accessed November 2018). As an example, recent studies of colorectal cancer (CRC) have demonstrated that careful and effective patient selection can lead to many useful conclusions for both clinical trials and therapeutic approaches.2 3 It is noteworthy that personal demographic characteristics have an effect on participation in biomedical research. The continuous generation of scientific information makes it difficult to organise a particular patient recruitment system (PRS).4 Moreover, a combination of reasons, such as the frequent diversity of hospital information systems (HIS) and electronic health records in health units, the daily busy clinical practice and also individuals’ personal barriers, impedes the creation of a fully unified PRS of a particular disease.5 6 Attempts to design recruitment strategies focusing on participants’ awareness and profiles have been previously published.7–11
As informatics becomes increasingly important in all fields of research, biomedical scientists must collaborate with computer professionals. The contribution of health informatics and biomedical technology has become more evident than ever before within the global scientific community, part of which involves the creation and exploitation of large data repositories.12 Most HIS maintain patients’ information in their internal network environment, making it difficult to collect and share data. On the other hand, national registries have immense potential for regulation of healthcare management and interdisciplinary development at a national level.13
Typically, patient registries constitute key ways to pool data. After a long period of clinical, molecular or genetic data collection, a patient database can be created from the recognised state, a private laboratory or other health agencies. On the other hand, this approach can sometimes hinder the dissemination of information. Such a strategy ultimately makes it difficult for the researcher to review the clinical and molecular profile of a patient thoroughly, in order to match eligible participants for a clinical trial. From this standpoint and with the motivation for national healthcare improvements in Greece, new eGovernment policies could be taken. Generally, in Greece, there are few patient centred registries, and many are not known to the scientific community. Additionally, none has unambiguous registration forms. Table 1 presents the PubMed database survey results related to Greek registries.
PubMed database search results related to Greek registries
In the present work, we introduce a virtual approach to a national biomedical registry of available patients for clinical trials in Greece, combining patient clinical and molecular profiles with Internet technology. We named the demonstration system the Hellenic Biomedical Registry (HBR). HBR as a proposed eGovernment policy is designed to help physicians of a health unit to better organise and run clinical studies by evaluating biomedical records through a web based framework. Furthermore, the registered physicians (MDs) are intended to function as data sources of our registry. Any patient who receives treatments in the healthcare system for his/her condition is potentially a data source for a physician to store the health case in a registry.
We chose CRC as an example for our study. CRC remains one of the most common and studied cancer types in both men and women, accounting for 862 000 deaths in 2018 worldwide, according to the WHO (http://www.who.int/mediacentre/factsheets/fs297/en/, last accessed December 2018). Its incidence and mortality rates vary by race and ethnicity without ignoring other factors, such as access to health services, modern anxiety lifestyle, increased obesity and lack of exercise.14 Moreover, the classification and molecular genetics of CRC could be used as an example for the integration of biomedical data and Internet network potential as an enhancement strategy of clinical trials.15–17
The article aims to prove that the HBR as a local host implemented system is applicable to any higher level healthcare manager, such as the Ministry of Health, with the objective of enhancing clinical trial intentions and processes.
Methods
We implemented our virtual web based software using the intranet of the Biomedical Engineering Laboratory, National Technical University of Athens, School of Electrical and Computer Engineering. The source code and the database schema are available on request. Our assumptions involved the following: (a) exploitation of a computer local host server as a development environment; (b) the need for minimal development cost; (c) and consideration that all physicians already have a index of digital literacy in the modern era. Our methodology involved four stages: required data elements; system development environment; system deployment; and datasets and system virtual implementation. A literature review was conducted for advisory related work and key points that concerned our approach.18–33 More specifically, the literature review aimed at: (a) the design of an integrated virtual framework; (b) evaluation of previous registry applications; (c) a search for minimal cost strategies for developing a modern web based platform involving conditions of interoperability; and (d) the need to integrate clinical and genomic data, using CRC as an example application for our framework. The next sections describe the methodology followed.
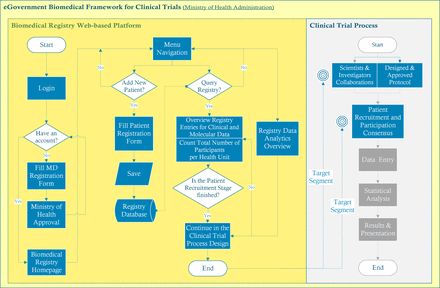
Basic system flowchart
The basic flowchart represents the main algorithmic steps consisting of object oriented programming modules. Figure 1 demonstrates a condensed visual approach of the system design, which was convincingly demonstrated to be effective in mapping the initial stage of our methodology.
(A) Virtual biomedical registry workflow. The proposed web based framework involves displaying the total number of patients registered for clinical trial participation per International Classification of Diseases, 10th edition (ICD-10) condition and per health unit. (B) Proposed Hellenic Biomedical Registry framework involves the initial segments of the clinical trial process.
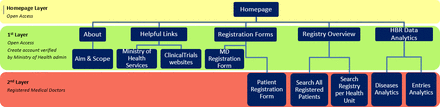
Registry interface structure
The user interface was designed to be organised into categories to be as user friendly as possible. As figure 2 shows, on the homepage and first layer of access, the physician of each health unit should be registered in the web platform by submitting the necessary MD registration form. It is assumed that the Ministry of Health should approve the physician’s application by sending credentials for the HBR using his/her personal email. Using his/her credentials, the physician gains access to the second and most important layer of the HBR platform and he/she is able to submit his/her patient's information and upload his/her consent to participate in the design of clinical trials related to his/her condition. The physician at that level has the ability to search patient profiles per health unit and per ICD-10 in which he/she is interested, which is the most important objective of our framework.
Hellenic Biomedical Registry (HBR) website navigation.
Required data elements
Ιn practice, it is generally not possible for a registry to collect and store all of the desired data.34 Therefore, using CRC as an example, we identified CRC centred clinical trials by searching the https://clinicaltrials.gov database to streamline the patient data elements (DEs) that researchers seem to consider important in order to complete registration forms on a patient recruitment system during the clinical trial design process.
Text mining of the extracted database information with the help of the RapidMiner tool35 had the following objectives: (1) to define key points in the inclusion/exclusion/intervention criteria; (2) to convert the aggregated DEs into appropriate fields and types including them into the database schema for our system; and (3) to adapt current recruitment processes to our own ideas for national biomedical registries.
All CRC DEs were designed to be stored in an online case report form (CRF). The CRF contains DEs that help researchers in organising a clinical study, but also in the calculation of statistics. It represents the patient’s registration form, completion of which is the responsibility of the registered physician, and it has a significant role in determining the quality and functionality of a registry.36 Unstructured textual DEs in the CRF also need to be included. We searched the PubMed database for review articles on the existing field of the molecular and genetic basis of sporadic and hereditary CRC to include clustered CRC information as molecular DEs in the framework design. The studies were limited to those published within the p[ast 5 years to ensure a modern perspective.
The above approach concerning DEs led to the creation of a biomedical record based on four categories: general and patient demographic data, and patient disease data (clinical and molecular/genomic data). Table 2 shows the DEs required to be submitted into the registry by completing specific data fields. For the patient’s molecular/genomic profile in particular, we organised the potential molecular diagnostic results by gene panels. Thus the physician can choose the genes examined based on the CRC panel test, recording in the registry important information about the patient’s molecular diagnostic results.
Data elements required to be submitted in the registry
It was necessary to establish the validity of our research by demonstrating that our virtual biomedical web application is efficient. Thus we selected a large tertiary hospital in Athens (251 General Hospital of the Hellenic Air Force, https://www.haf.gr/structure/gea-2/251gna/, last accessed December 2018), which employs approximately 2000 workers and serves more than 250 000 beneficiaries of hospitalisation per year, in order to collect real patient data for a reliable simulation of the application’s functionality. In more detail, we queried the database of the hospital information system for the total number of distinct patients who were hospitalised for a minimum of 1 day during 2016 and 2017. Approval of database queries was obtained from the research, ethics and education committee of the hospital. The new GDPR (https://gdpr-info.eu/, last accessed December 2018) regulation was also considered in order to ensure the highest privacy setting by anonymisation of the data collected. In the sequel, data were filtered for CRC incidents and divided into two sets. The total number of distinct patients who were hospitalised in 2016 are referenced in our application as 'health unit A', and all corresponding patients of 2017 are referenced as 'health unit B'. Hence we briefly simulated real patient datasets from two different stochastic hospitals, ‘health unit A’ and ‘health unit B’. The datasets were imported into the MySQL database structure of the application and we eventually implemented the full system to a local host environment of our laboratory intranet. Dashboard administration allowed a modern user experience and user interface composition that permitted a grid view appearance of our structured database queries and the application algorithm.
From a health information security perspective, it must be emphasised that the personal data of the patients should always be anonymised. From data holders such as the Ministries of Health to software developers, security issues related to controlling and sharing health data must receive significant attention. Data anonymisation techniques related to logical and physical security perspectives are outside of the scope of our system demonstration at this time. However, we implemented the minimum required processes with respect to security issues: (1) our registry is designed to be accessible by credentials; (2) we replaced each patient’s health unit ID from our real datasets with an HBR ID (HBRID) in the database back end environment; and (3) a WordPress open source plugin for software GDPR compliance was included in our source code. Finally, from a network architecture perspective, it is anticipated that our proposed framework could work under a secure virtual private network (VPN) of the Ministry of Health managing authority. A proposed security policy is analysed in the Supplementary File.
Supplemental material
Results
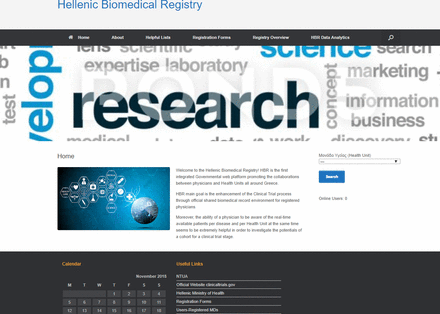
We present an effective virtual prototype biomedical registry implementing the idea of real time evaluation of available registered patients nationwide, querying their clinical and molecular profiles as an enhancement strategy for clinical trials in Greece. Our virtual web application runs through any browser on a local host web server. Figure 3 shows the screenshot of the landing page. This system is designed to be hosted and managed by the Officials of the Hellenic Ministry of Health. As figure 1B shows, the proposed web based framework can improve collaborations among researchers for an effectively designed clinical trial protocol. Our national biomedical registry might serve as: (1) a highly effective PRS including comprehensive information about patient participation and (2) an intelligent decision support system for evaluating the potential of hospitalised patients or outpatients to participate in clinical trials.
Screenshot of the landing page of the Hellenic Biomedical Registry running on a local host web server (http://localhost/wordpress/home).
Registry case report form
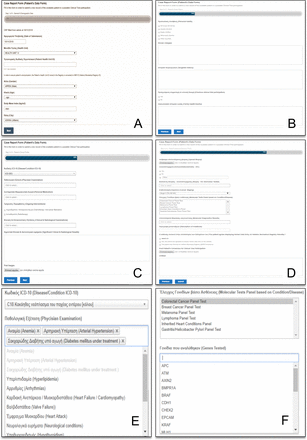
Through the virtual HBR, any registered physician can submit a patient’s clinical and molecular/gene data together with that patient’s participation consent form in a clinical trial. All necessary DEs are stored through patient centred web forms, creating a dynamic biomedical record. Figure 4 shows screenshots of the four sequential web pages that compose the CRF in the demonstration system according to table 2. The data fields of the CRF consist of text boxes, dropdown lists, multiselect fields and advanced file upload buttons. Clustering of CRC molecular diagnostic information helps the user to easily locate the gene panel that has been tested in a patient. Storage of pathological and mutant genomic information automatically creates conditions for easier selection of patients with similar gene profiles and facilitates comparisons with their clinical status or treatment. It should be mentioned that the storage of information on genomic mutations should follow the internationally recognised nomenclature.
Screenshot examples of the patient registration form that is referred to as the case report form (CRF); it is based on four sequential web pages. Completion of the CRF is indicated by a progress bar. All steps are completed by the physician. On the first CRF web page, the physician can submit the patient’s general and demographic data (A). On the next two web pages, the physician can store data about the patient’s clinical condition and radiological tests (B, C). The final CRF web page stores information about the patient’s biopsy, immunohistochemistry and molecular diagnostic results (D). (Ε) Multiselect option for the radiological examinations completed by the patient. (F) Molecular diagnostic tests in the form of gene panels in which the patient has been tested. The multiselect dropdown list of genes is enabled logically when the corresponding panel test is selected.
Overview of available patients for clinical trials
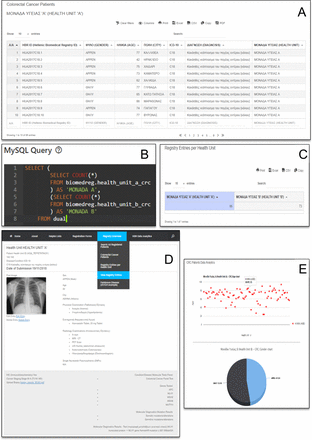
Each registered MD can overview all the participants per health unit. Figure 5 displays screenshots reflecting our biomedical registry philosophy. In more detail, filtering for CRC patients in ‘health unit A’, the MD user sees the grid view of image figure 5(A). For a more comprehensive overview of the available patients associated with a certain disease/condition per health unit, the HBR runs 'select count' SQL commands comparing two or more health units’ data. Running the 'select count' SQL command with our realistic simulated patient data that were divided into two virtual datasets, we find the information that the health units ‘A’ and ‘B’ maintain a significant number of CRC prospective participants. Displaying such information, an MD user effectively has a complete set of patients with a specific disease. This is the first step before further filtering patients to join a clinical trial.
Screenshot examples of the virtual Hellenic Biomedical Registry (HBR) implementation overview based on real patient data. (A) Colorectal cancer (CRC) patients of the virtual health unit ‘A’. Executing the 'select count' SQL command for only two health units, we generate the numerical comparison of available patients with the same International Classification of Diseases, 10th edition (ICD-10) code (B, C). Each set of case report form information stored in the registry is displayed as the corresponding patient’s biomedical record, which eventually can be accessed by other registered physicians (D). Example of HBR data analytics. (E) Bubble chart represents the CRC patients of ‘health unit A’ characterised by their age. Hovering over a dot, the physician is informed of the patient’s age and gender. From another aspect, the physician can use the chart to assess an overview of the age range of patients with a particular disease in a health unit. Τhe pie chart shows the percentage of male and female CRC patients in ‘health unit B’.
Overview of patient’s biomedical record
A completed CRF is basically extended to a corresponding patient biomedical record. Figure 5(D) displays the CRF data entry of a CRC patient. All of the information in each CRF entry is presented in a sequential order, providing a single patient’s comprehensive biomedical record. The uploaded files from the CRF step are available to be downloaded and evaluated by the registered MDs. Through the platform menu, each MD user can review the biomedical information of a patient in a health unit. This is helpful for an MD of another health unit to review any condensed patient medical records attended by a colleague. Moreover, this ability can promote collaboration between MDs of different health units and act as an incentive to carry out clinical research.
Data analytics contribution
In healthcare, data analytics are used to enable stakeholders to make informed decisions.37 Healthcare registries are an important source of information associated with patient outcomes. Hence HBR emphasises data entry visualisation, providing valuable insight into the general and more specific profiles of participants. Several types of plots, including pie charts, histograms, bubble charts and line charts, are used to visualise HBR patients outcomes. figure 5(E) shows an example of HBR data analytics. Overviewing data analytics, registered physicians may decide whether they have the eligible cohort of patients to initiate a clinical trial.
Discussion
Clinical trials generate a vast amount of data. Clinical trial stakeholders (ie, sponsors, governmental services, research institutes, universities, biomedical investigators) should foster a culture in which data sharing is the expected norm.38 39 Following this objective, implementation of the biomedical registry in Greece aims at enhancing the planning of clinical trials and further health management. The proposed system assists physicians in data analysis, and limits the time needed to find and submit eligible patients for a clinical trial by displaying integrated key data elements (demographics, clinical, molecular information). By importing accurate data, it can be considered as an intelligent decision support system with clinical and molecular query potentials. However, the data displayed in such a proposed web based system must always be valid. Querying prospective patients to participate in a clinical trial requires that the associated data must be accurate at all times. Hence the physicians as the data sources of the registry must be very careful in selecting the appropriate patients for submission and in updating patient biomedical information, and should be GDPR compliant. The likelihood of survival during a clinical trial process as well as the ongoing supervision of the patient by the same physician should be a matter of serious consideration when recruiting patients.
Another point to consider is that our web based biomedical registry can also support the recording of patients with rare diseases. However, this could be achieved on the condition that the philosophy of our registry would fit into a more general eGovernment health framework as the healthcare information systems of our time are moving from autonomous technologies to more interoperable ones. Therefore, a careful and forward looking eGovernment policy on health registry design could solve important issues in the fields of patient recruitment systems, clinical trial processes and eventually personalised medicine. For example, instead of relying on the developer's imagination by creating more registries or unpublished databases, each DE that pertains to a patient's biomedical information could be standardised, forcing each registry to use the same DE type.40 We believe that the adoption of a web based modular system architecture combined with integration of clinical and molecular information, such as our HBR, would encapsulate each autonomous registry as part of a national health information superset.
Lastly, it is necessary that if a governmental agency chooses to adopt our idea, penetration testing procedures will need to be thoroughly executed, and information security policies will need to be updated prior to software implementation.
Conclusion
In contrast with the approaches of announcing and serving data of designed, ongoing and upcoming clinical trials, we describe a modern dynamic web based modular with extremely low development costs. The use of free, open source software enables the high level customisation of our national biomedical registry. However, it should be emphasised that the technical design details of the application, as well as data security and implementation issues, have not been the focuses when designing this system. What we think is important is the motivation behind the work. Such a biomedical registry has never been established or managed as part of the Hellenic eGovernment. Extensive research on the Internet has allowed us to confirm that such a framework has never been proposed at a national level. The combination of real web development and virtual demonstration of a system that exploits realistic patient data, implies the potential substantial usability of our registry for the enhancement of clinical trial processes.
In conclusion, the designed Hellenic Biomedical Registry manages specific data structures and relationships, aiming to provide four novel approaches under one philosophy:
Direct ability of a physician to import his/her patient’s data into a registry, empowering participation in clinical trials.
Integration of clinical and molecular/genomic data under a national biomedical registry.
Web monitoring and evaluation of nationally available patients per disease/condition and per health unit for the design of a clinical trial or for the selection of an eligible patient cohort.
Creating for physicians a review framework of the incidents of their medical specialty that are being hospitalised per health unit, promoting a variety of medical approaches, collaborative research and the ability to reach wider populations.
References
Footnotes
Contributors AK and D-DK conceived the idea and developed the theory. AK designed the model, wrote the source code and gathered the appropriate data. GL supervised the implementation stage and verified the simulation results. AK and GL wrote the manuscript. All authors discussed the results. D-DK directed the project.
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests None declared.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; internally peer reviewed.